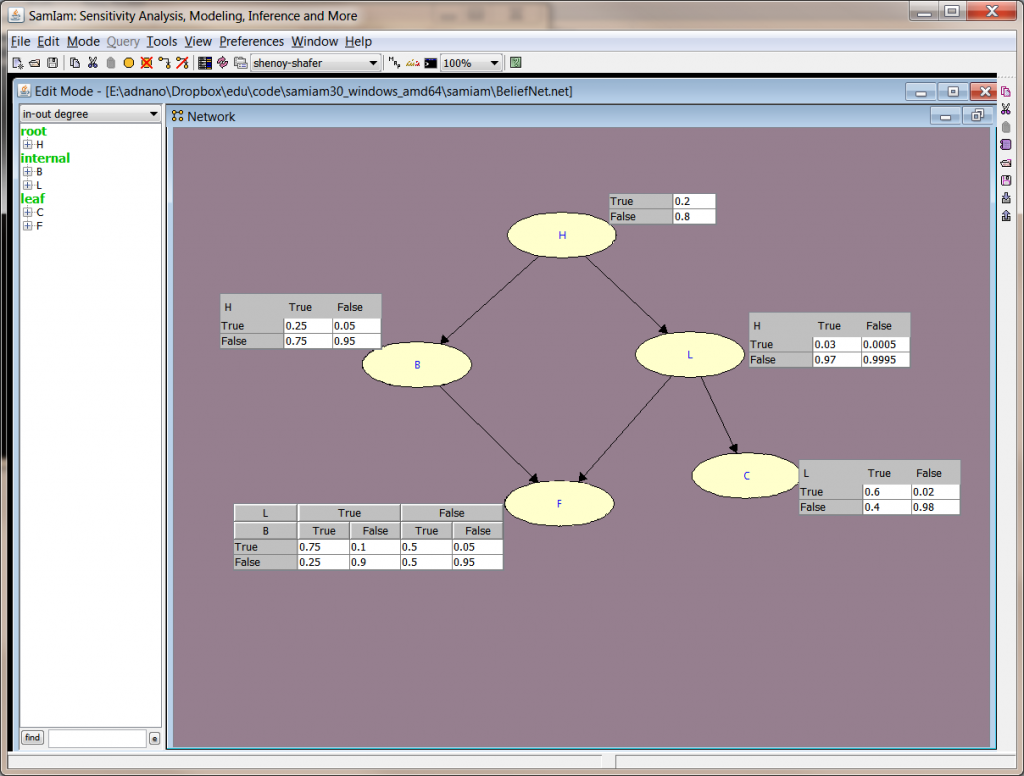
As discussed previous post on Customizing Conditional Probability using Code Generation with SamIam, I have touched upon importance of having programmatic and declarative control over the network. Working with SamIam (and with Infer.NET to some extent) gives a researcher provides this flexibility which is hard to find in proprietary tools.
Here is a simple example of a typical text book belief network. Once graphically drawn, SamIam's code bandit allow you to extract the model out as a class.
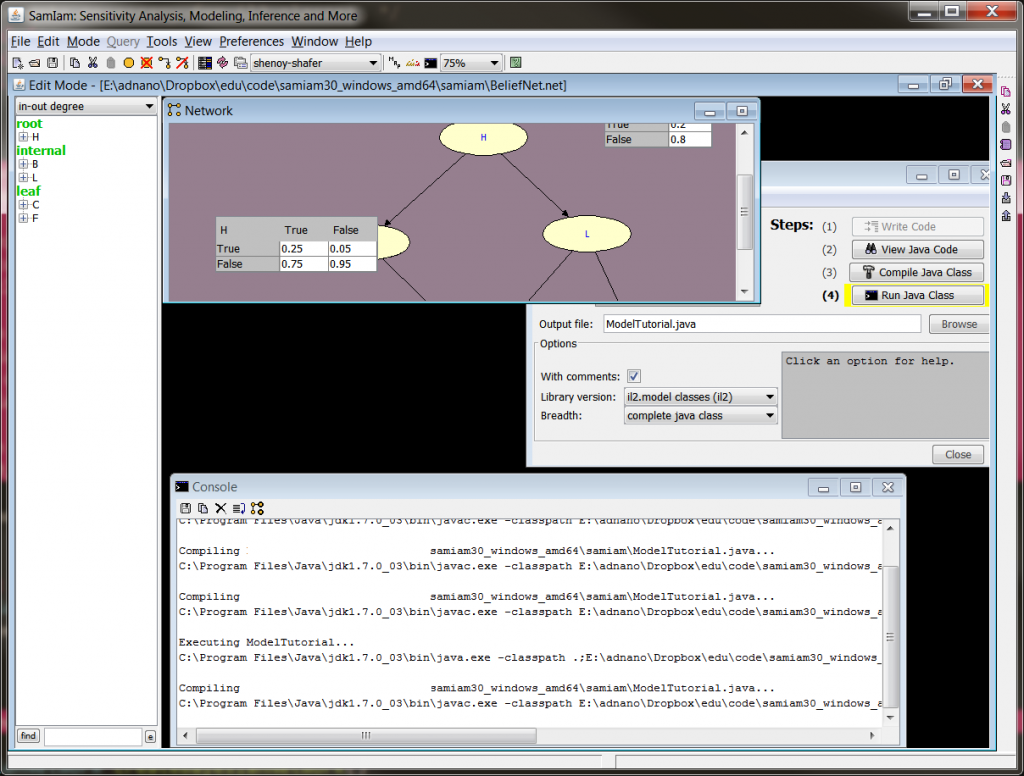
This class hard codes the network ...code\samiam30_windows_amd64\samiam\BeliefNet.net where one can operate on the object BayesianNetwork and can modify the nodes by population from a different data source rather than hard coding. Once a simple, readable structure model is available in raw code, there are lots of possibilities for data population. To build, ensure that inflib.jar occurs in the command line classpath, e.g. javac -classpath inflib.jar ModelTutorial.java
public BayesianNetwork createBayesianNetwork()
{
/* Create a domain of size 5. */
Domain domain = new Domain(5);
/* Add a discrete variable called "H" to the domain,
with states "True", "False". */
String name0 = "H";
String[] values0 = new String[]{ "True", "False" };
int id0 = domain.addDim( name0, values0 );
/* Add a discrete variable called "B" to the domain,
with states "True", "False". */
String name1 = "B";
String[] values1 = new String[]{ "True", "False" };
int id1 = domain.addDim( name1, values1 );
/* Add a discrete variable called "L" to the domain,
with states "True", "False". */
String name2 = "L";
String[] values2 = new String[]{ "True", "False" };
int id2 = domain.addDim( name2, values2 );
/* Add a discrete variable called "C" to the domain,
with states "True", "False". */
String name3 = "C";
String[] values3 = new String[]{ "True", "False" };
int id3 = domain.addDim( name3, values3 );
/* Add a discrete variable called "F" to the domain,
with states "True", "False". */
String name4 = "F";
String[] values4 = new String[]{ "True", "False" };
int id4 = domain.addDim( name4, values4 );
/* For the cpts, create arrays of double-precision floating point values. */
//H Value
//True 0.2
//False 0.8
double[] cpt0 = new double[]{ 0.2, 0.8 };
//B H Value
//True True 0.25
//True False 0.05
//False True 0.75
//False False 0.95
double[] cpt1 = new double[]{ 0.25, 0.05, 0.75, 0.95 };
//L H Value
//True True 0.03
//True False 5.0E-4
//False True 0.97
//False False 0.9995
double[] cpt2 = new double[]{ 0.03, 5.0E-4, 0.97, 0.9995 };
//C L Value
//True True 0.6
//True False 0.02
//False True 0.4
//False False 0.98
double[] cpt3 = new double[]{ 0.6, 0.02, 0.4, 0.98 };
//F L B Value
//True True True 0.75
//True True False 0.1
//True False True 0.5
//True False False 0.05
//False True True 0.25
//False True False 0.9
//False False True 0.5
//False False False 0.95
double[] cpt4 = new double[]{ 0.75, 0.1, 0.5, 0.05, 0.25, 0.9, 0.5, 0.95 };
Later on, SamIam creates the table using the CPT's and eventually build the network using these tables.
/*
Create a IL2 Table for each cpt.
The parameters to the Table constructor are:
(1) the domain,
(2) the variable ids that name the dimensions of the table (in the form of an IntSet),
(3) the cpt data.
*/
Table table0 = new Table( domain, new IntSet( new int[]{ id0 } ), cpt0 );
Table table1 = new Table( domain, new IntSet( new int[]{ id0, id1 } ), cpt1 );
Table table2 = new Table( domain, new IntSet( new int[]{ id0, id2 } ), cpt2 );
Table table3 = new Table( domain, new IntSet( new int[]{ id2, id3 } ), cpt3 );
Table table4 = new Table( domain, new IntSet( new int[]{ id1, id2, id4 } ), cpt4 );
/* Create an array of all the Tables. */
Table[] tables = new Table[]{ table0, table1, table2, table3, table4 };
/*
The simple BayesianNetwork constructor takes only one argument:
an array of Tables.
*/
BayesianNetwork model = new BayesianNetwork( tables );
Upon building, you get the following console output.
Happy inferring!